Richard Sternberg is an evolutionary biologist with interests in the relation between genes and morphological homologies, and the nature of genomic “information.” He holds two Ph.D.'s: one in Biology (Molecular Evolution) from Florida International University and another in Systems Science (Theoretical Biology) from Binghamton University. From 2001-2007, he served as a staff scientist at the National Center for Biotechnology Information, and from 2001-2007 was a Research Associate at the Smithsonian’s National Museum of Natural History. Dr. Sternberg is presently a research scientist at the Biologic Institute, supported by a research fellowship from the Center for Science and Culture at Discovery Institute. He is also a Research Collaborator at the National Museum of Natural History.
From 2001-2004, Sternberg served as Managing Editor of the Proceedings of the Biological Society of Washington, and has also served on the Editorial Board of the International Journal of General Systems. In 1999, he was a Visiting Associate Professor of Biology at Northern Michigan University, and from 1999-2001 was a Postdoctoral Fellow in the Department of Invertebrate Zoology, National Museum of Natural History. He has received postdoctoral fellowships from both the NIH and the National Museum of Natural History, and has published refereed articles in such journals as Genetica, Evolutionary Theory, Journal of Comparative Biology, Crustacean Research, Journal of Natural History, Journal of Morphology, Journal of Biological Systems, and the Annals of the New York Academy of Sciences.
For a more detailed discussion of his approach to biology, see his essay, "How My Views on Evolution Evolved." For information about Dr. Sternberg's mistreatment by the Smithsonian Institution due to his sympathy for intelligent design, see the Smithsonian Controversy page from his personal website, RichardSternberg.com.
Archives

DNA May Be “Junk” at One Level But of Utmost Significance at Another
Chromosome Dynamics Has Egg-centric Features

The “Why” of the Fly “Y”: Reflections on “Junk” DNA
Richard Sternberg on the Trail of the Immaterial Genome

Dr. Richard Sternberg: Whale Evolution and Living Waters, Pt. 2

Dr. Richard Sternberg: Whale Evolution and Living Waters

On Human Origins: Is Our Genome Full of Junk DNA? Pt 5

On Human Origins: Is Our Genome Full of Junk DNA? Pt 4
On Human Origins: Is Our Genome Full of Junk DNA? Pt 3
On Human Origins: Is Our Genome Full of Junk DNA? Pt 2.

On Human Origins: Is Our Genome Full of Junk DNA?

Limited-Time Offer! Act Now! Operators Standing By! MagiMold™!

Evolutionary Biologist Richard Sternberg discusses modern genomics and junk DNA
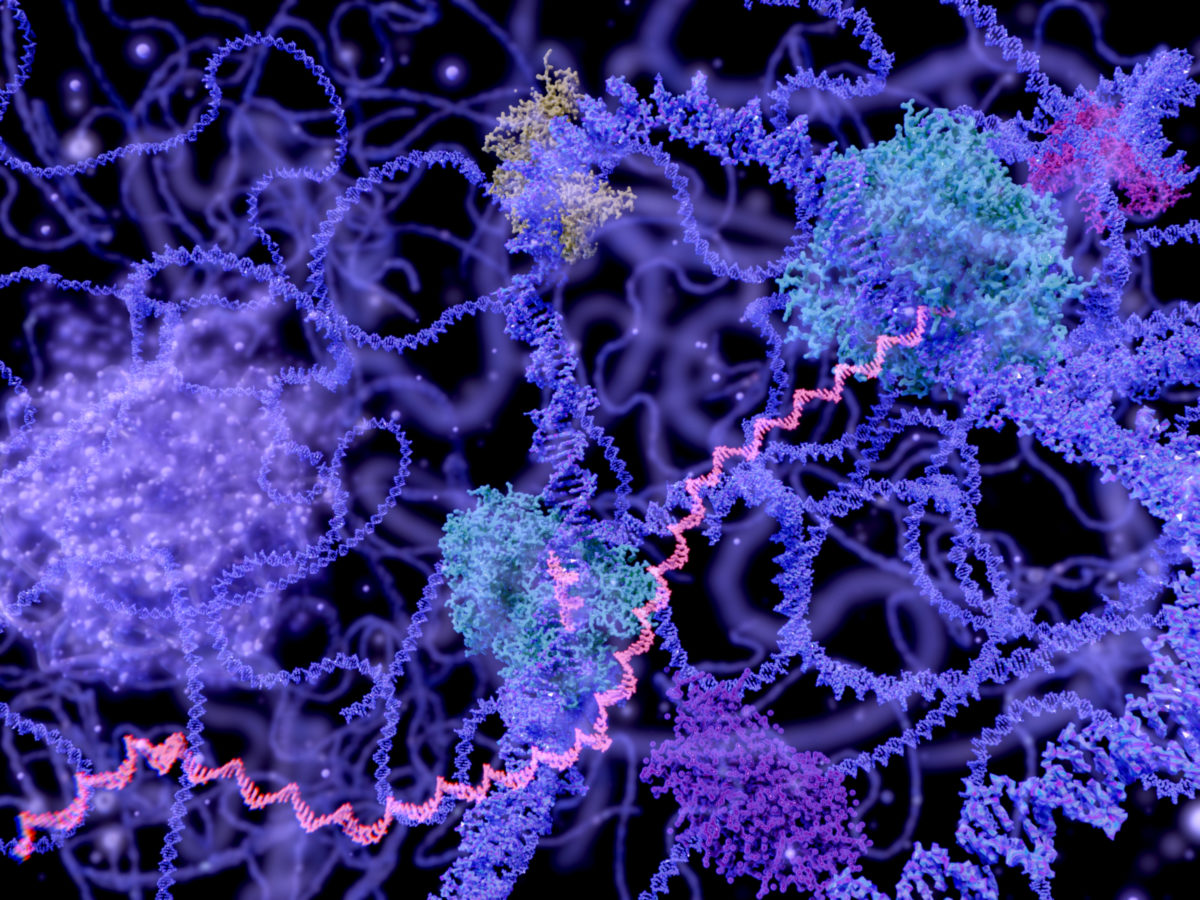
Matheson’s Intron Fairy Tale

Let’s Do the Math Again
Beginning to Decipher the SINE Signal
Discovering Signs in the Genome by Thinking Outside the BioLogos Box