Responding to Criticisms of Irreducible Complexity of the Bacterial Flagellum from the Australian Broadcasting Network
Time flies. Published in 1996, Michael Behe’s book Darwin’s Black Box represented a major development in the modern articulation of the scientific argument for intelligent design. The book will have its 20th anniversary in 2016. Behe was in town for our Summer Seminar on Intelligent Design, and we were talking about this — and about how despite the passage of time, critics still object ineffectually to his argument based on irreducible complexity (IC). They make the same arguments, which still fall flat for the identical reasons they did years ago. We’ve documented this repeatedly. Yet they keep coming at us.
In July, 2015, an article turned up at the Australian Broadcasting Corporation, “The bacterial flagellar motor: brilliant evolution or intelligent design?,” by a biophysicist named Matt Baker, claiming to refute irreducible complexity. Has Baker finally solved the riddle of answering Behe’s challenge to Darwinian evolution? No, it’s pretty much the same stuff we’ve heard before, with maybe a variation or two that are original to Matt Baker. Well, since these perennial objections are indeed perennial, I would like to answer them again, using Dr. Baker as my example. We’ll see that IC remains as potent a weapon in ID’s arsenal as it was in 1996.
The article purports to explain how the bacterial flagellum is the result of Darwinian evolution rather than intelligent design. But the author badly misunderstands both how we test for irreducible complexity and what it means to provide a Darwinian explanation. He is also apparently unaware of the many reasons why the Type III Secretory System could not have been a precursor to the flagellum.
Part 1: Addressing Matt Baker’s Misunderstandings About How We Test Irreducible Complexity
The article’s first error comes in the sub-headline, which states:
Luckily, individual components of the bacterial flagellar motor have indeed been found elsewhere. And they work. So the motor is ‘reducible’, and certainly not ‘irreducibly complex’.
First of all, it’s not the case that all “individual components” of the flagellum have been found elsewhere. But even if they had, that would not necessarily mean that the motor is “reducible” and “not ‘irreducibly complex.'” In any case, Baker goes on to state:
A central tenet of this theory is the notion of ‘irreducible complexity’. This asserts that some biological machines — like the flagellar motor — must be the product of design, because if you were to remove one or two components from the motor it would not function properly, or at all. The logic being, this motor was designed as a whole construction — it didn’t evolve through a series of steps, so the individual parts of the motor would serve no purpose on their own.
So the creationist argument relies on us finding no evidence of individual parts of the motor having a role outside of bacterial flagella.
Ignoring the gratuitous “creationist” jab, his argument is self-contradictory. On the one hand he says (correctly) that irreducible complexity means that a system “didn’t evolve through a series of steps.” But he then wrongly claims that this implies “the individual parts of the motor would serve no purpose on their own” or that irreducible complexity “relies on us finding no evidence of individual parts of the motor having a role outside.” The former claim is a great description of irreducible complexity; the latter is a straw man test, which has nothing to do whatsoever with the concept.
Dr. Baker should read my article “Do Car Engines Run on Lugnuts? A Response to Ken Miller & Judge Jones’s Straw Tests of Irreducible Complexity for the Bacterial Flagellum,” which addresses this common misconception. I explain there that Michael Behe formulates irreducible complexity as a test of building an entire system in a stepwise manner. IC relates to the functionality of a collection of parts, not the function (or possible functions) of each individual part. Even if a separate function could be found for a sub-system or sub-part, that would not refute the irreducible complexity of the whole, nor would it demonstrate the evolvability of that entire system. Here’s how Behe defines IC:
In The Origin of Species Darwin stated:
“If it could be demonstrated that any complex organ existed which could not possibly have been formed by numerous, successive, slight modifications, my theory would absolutely break down.”
A system which meets Darwin’s criterion is one which exhibits irreducible complexity. By irreducible complexity I mean a single system composed of several well-matched, interacting parts that contribute to the basic function, wherein the removal of any one of the parts causes the system to effectively cease functioning.
Michael Behe, Darwin’s Black Box, pg. 39 (Free Press, 1996).
According to Darwin himself, Darwinian evolution requires that a system be functional along each small step of its evolution. One could find a sub-part that could be useful outside of the final system, yet the total system would still face many points over its “evolutionary pathway” where it could not remain functional through “numerous, successive, slight modifications.” Thus, Baker mischaracterizes Behe’s argument as one that focuses on the non-functionality of sub-parts, when in fact, Behe actually focuses on the ability of the entire system to assemble in a stepwise fashion, even if sub-parts can have functions outside of the final system.
To further understand how Baker’s test fails, consider the example of a car engine with its nuts and bolts. Car engines use many kinds of bolts, and a nut or a bolt could be seen as a small “sub-part” or “sub-system” of a car engine. Under this logic, if a vital nut in my car’s engine might also perform some other function — perhaps as a lug nut — then it follows that my car’s whole engine system is not irreducibly complex. Such an argument is obviously fallacious.
In assessing whether an engine is irreducibly complex, one must focus on the function of the engine itself and whether it can be built in a stepwise fashion, not on a possible function that one particular sub-part could have elsewhere. Of course a nut or bolt could serve some other purpose in my car. It could probably serve many purposes. But this does not explain how a variety of complex parts such as pistons, cylinders, the camshaft, valves, the crankshaft, sparkplugs, the distributor cap, and wiring came together in the appropriate configuration to make a functional engine. Even if all of these parts could perform other functions in the car (which is doubtful), how were they all assembled properly to construct a functional engine? The answer must be intelligent design.
To offer another analogy, consider how you would build an irreducibly complex arch (Figure A):
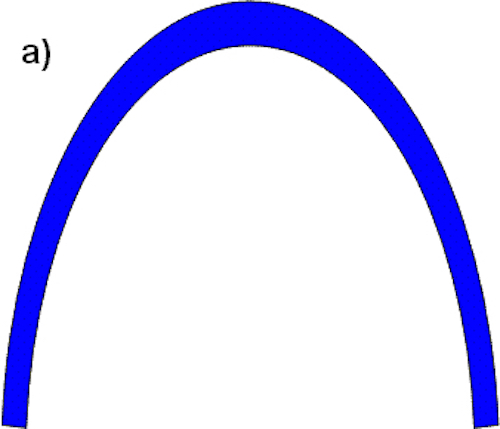
According to Baker, if we can find a function for some sub-piece, then a system is not irreducibly complex. Now, let’s now break this arch into sub-pieces:
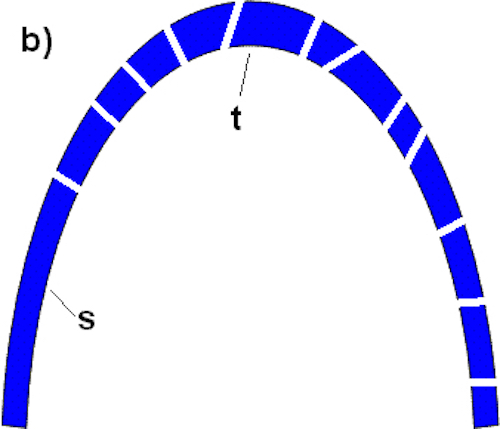
Baker has apparently found a flagellar sub-piece (the T3SS) that can perform some other function. The T3SS comprises no more than a quarter of the total flagellar parts. Similarly, in this arch, there is one large sub-section (labeled “S”) that comprises approximately a quarter of the total arch. Sub-section S can have a function outside of the arch (i.e., it can stand on its own). However, this exposes the fallacy of Baker’s test: the ability of sub-section S to stand on its own does not therefore dictate that the arch is not irreducibly complex. If one were to remove the top piece (t), the arch crumbles, even if sub-section S remains standing (Figure C):
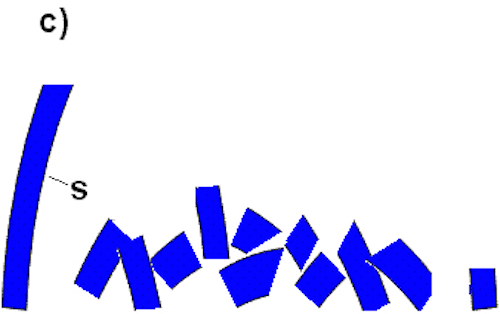
Thus, we see that a system does not become “reducibly complex” simply because one part remains functional outside of the final system, and Baker has followed many others in proffering a straw-man test of irreducible complexity.
So can we properly test the flagellum to show that it is irreducibly complex? Yes, we can. Scott Minnich’s genetic knockout experiments on the E. coli flagellum have shown that it fails to assemble or function properly if any one of its approximately 35 structural parts are missing. That’s prima facie evidence that it’s irreducibly complex, and it’s a proper test of the model.
Part 2: Why the Type III Secretory System Can’t Be a Precursor to the Bacteria Flagellum
In his article, Dr. Baker thinks he’s found one main flagellar component that can function outside the flagellar system and that this refutes irreducible complexity. It’s nothing new to ID proponents. It’s called the Type III Secretory System (T3SS) and it’s basically a pump that’s used to transport proteins across the cell membrane of bacteria. Baker frames his case this way:
Proof of the flagellar motor’s ‘reducibility’ — that its component parts can function elsewhere — comes in the form of the injectisome; another fabulous molecular machine found in bacteria. This needle-like complex is used by disease-causing bacteria to punch holes in the host’s target cells.
The protein machinery used to assemble the proteins that make up the punching needle is identical to that used to assemble the ‘propeller’ part of the flagellar motor — the filament and hook of the motor. In addition, nine core proteins of the flagellar motor share common ancestry with injectisome proteins — the genes that code for them are so similar they have clearly come from the same genetic ancestor.
This argument is so old that it was already dealt with and refuted in the 2003 ID documentary Unlocking the Mystery of Life.
So in the interest of accuracy, let’s reframe Baker’s argument for him. Part of the flagellum is a molecular pump, the T3SS, which is used to pump proteins from inside the cell to the outside of the cell where they self-assemble into the flagellum. The injectisome uses the T3SS for a similar function — it is involved in both assembling the injectisome and in the injectisome’s function.
The injectisome is used by certain predatory bacteria to inject toxic proteins into eukaryotic cells, which then kill the eukaryotic cells so they can be ingested by the bacterium. It’s just a molecular pump; that’s it.
Baker suggests that the injectisome’s proteins are “identical” to the “flagellar motor” but the T3SS isn’t actually part of the motor portion of the flagellum. It’s only part of the pump that is used during flagellar assembley, which then also serves as the basal body that anchors the flagellum in the cell membrane.
Nor is it the case that these proteins are “identical.” I’m quite willing to grant that some proteins between the T3SS and the flagellum appear homologous, but it’s certainly not correct to call them “identical.” One authoritative paper by Pallen and others in 2005 suggests that a flagellar protein (FliK) and a protein from the T3SS (YscP) are “homologous” because, across a stretch of about 58 amino acids within their 300 or so amino acids, 13 residues are identical in at least 50 percent of the proteins studied, and another 23 amino acids within that stretch have similar chemical properties. (See Pallen et al. (2005), Figure 4.) That means that less than 25 percent of the amino acids are identical across a stretch that represents less than 20 percent of the total protein. While the proteins are clearly similar — perhaps very similar — and probably homologous, it’s misleading to say they are “identical.”
But let’s say they are identical — it doesn’t really matter. In fact let’s grant that nine of the proteins in the T3SS of the injectisome are identical to the proteins of the T3SS used by the flagellum. Does this constitute an evolutionary explanation? No, for many reasons.
First, at most the sequence similarity can establish, as Baker puts it, that “they have clearly come from the same genetic ancestor.” But Michael Behe is quick to remind us why establishing common ancestry is different from establishing a Darwinian explanation:
- “Although useful for determining lines of descent … comparing sequences cannot show how a complex biochemical system achieved its function — the question that most concerns us in this book. By way of analogy, the instruction manuals for two different models of computer put out by the same company might have many identical words, sentences, and even paragraphs, suggesting a common ancestry (perhaps the same author wrote both manuals), but comparing the sequences of letters in the instruction manuals will never tell us if a computer can be produced step-by-step starting from a typewriter. … Like the sequence analysts, I believe the evidence strongly supports common descent. But the root question remains unanswered: What has caused complex systems to form?” (Behe, Darwin’s Black Box, pp. 175-176.)
- “[M]odern Darwinists point to evidence of common descent and erroneously assume it to be evidence of the power of random mutation.” (Behe, The Edge of Evolution, p. 95.)
Second, it’s doubtful that the T3SS is useful at all in explaining the origin of the flagellum. The injectisome is found in a small subset of gram-negative bacteria that have a symbiotic or parasitic association with eukaryotes. Since eukaryotes evolved over a billion years after bacteria, this suggests that the injectisome arose after eukaryotes. However, flagella are found across the range of bacteria, and the need for chemotaxis and motility (i.e., using the flagellum to find food) precede the need for parasitism. In other words, we’d expect that the flagellum long predates the injectisome. And indeed, given the narrow distribution of injectisome-bearing bacteria, and the very wide distribution of bacteria with flagella, parsimony suggests the flagellum long predates injectisome rather than the reverse. As one paper observes:
Based on patchy taxonomic distribution of the T3SS compared to that of the flagellum, widespread in bacterial phyla, previous phylogenetic analyses proposed that T3SS derived from a flagellar ancestor and spread through lateral gene transfers.”
(Sophie S. Abby and Eduardo P.C. Rocha, “An Evolutionary Analysis of the Type III Secretion System” (2012).)
Likewise, New Scientist reported:
One fact in favour of the flagellum-first view is that bacteria would have needed propulsion before they needed T3SSs, which are used to attack cells that evolved later than bacteria. Also, flagella are found in a more diverse range of bacterial species than T3SSs. “The most parsimonious explanation is that the T3SS arose later,” says biochemist Howard Ochman at the University of Arizona in Tucson.
Now under normal evolutionary reasoning, one would take this kind of phylogenetic evidence to indicate that the flagellum long predates the T3SS, and that the T3SS is in no way a precursor (or closely related to a precursor) of the flagellum. But don’t expect evolutionists to use their normal reasoning when trying to oppose potent arguments for intelligent design. Here, they reject standard phylogenetic concepts like parsimony and assume that somehow the T3SS is (or is very similar to) some kind of a flagellar precursor. Normal evolutionary analysis would absolutely reject that hypothesis.
Additionally, it’s interesting that flagellar genes are always present (whether vestigial or suppressed) whenever injectisome genes are present. Thus, the injectisome is always found with the flagellum, but not vice versa. Again, this implies the injectisome has an origin that postdates the flagellum, and that it may even be derived from the flagellum.
So the evidence strongly suggests that the injectisome (or something like it) did not predate the flagellum, and thus can’t help explain how the flagellum evolved.
Finally, there’s the fact that the function of the T3SS (whether in the injectosome or in the flagellum) really has nothing to do with explaining the propulsion function of the flagellum. Again, the T3SS is just a pump that can move proteins across the cell membrane. It doesn’t in any way explain how the flagellum motor and its core propulsion function arose. I like how William Dembski captures the essence of the logical and empirical flaws in the T3SS argument:
[F]inding a subsystem of a functional system that performs some other function is hardly an argument for the original system evolving from that other system. One might just as well say that because the motor of a motorcycle can be used as a blender, therefore the [blender] motor evolved into the motorcycle. Perhaps, but not without intelligent design. Indeed, multipart, tightly integrated functional systems almost invariably contain multipart subsystems that serve some different function. At best the TTSS [Type III Secretory System] represents one possible step in the indirect Darwinian evolution of the bacterial flagellum. But that still wouldn’t constitute a solution to the evolution of the bacterial flagellum. What’s needed is a complete evolutionary path and not merely a possible oasis along the way. To claim otherwise is like saying we can travel by foot from Los Angeles to Tokyo because we’ve discovered the Hawaiian Islands. Evolutionary biology needs to do better than that.
William A. Dembski, Rebuttal to Reports by Opposing Expert Witnesses.
Given that Baker nowhere tries to provide any kind of a stepwise explanation for the evolution of the flagellum, he certainly has not met his standard of proof nor has he met Dembski’s challenge.
Part 3: Flagellar Diversity Challenges Darwinian Evolution, Not Intelligent Design
Dr. Baker has one other argument up his sleeve to try to show that the flagellum evolved. In his view, God wouldn’t have done it that way. Essentially, Baker has imbibed Nick Matzke and Mark Pallen’s fallacious theological argument that because there is diversity among flagella, the structure must not have been designed. He writes:
In the aftermath of the first legal challenges to curriculum requirements to teach intelligent design, evolutionary biologists Mark Pallen and Nicholas Matzke wrote “either there were thousands or millions of individual creation events … or one has to accept that the highly diverse contemporary flagellar systems have evolved from a common ancestor
Now maybe some (or even most) flagella are indeed related to one another, but there’s no reason why a designer couldn’t design diverse flagella. After all, in our experience with technological systems devised by human beings, we see incredible diversity! Look at all of the different ways that people have designed cell phones, trucks, or even something as simple as a key. These devices come in all kinds of myriad forms. Diversity doesn’t negate design.
Thus, Baker is proffering a theological argument that is simply irrelevant to the scientific case for intelligent design. In reality, flagella are distributed in a polyphyletic manner that doesn’t fit what we’d expect from common ancestry:
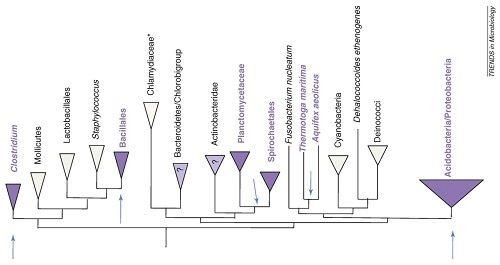
What you see in the figure above are various major groups of bacteria, represented by triangles (or in some cases written text). They are arranged here according to a standard phylogeny of bacteria. The purple groups have flagella throughout the clade, and the groups with question marks have only a minority of species with flagella within that clade. The white triangles show groups not thought to have flagella.
What’s the problem? The groups with flagella are scattered all about the tree and do not form a single monophyletic group. In other words, the diversity of flagella cannot be easily explained by common ancestry. Writing in Trends in Microbiology, the authors of the figure reprinted above explain the problem:
When we attempted to map the known distribution of flagellar genes on to a recently published ‘tree of life’, instead of a single monophyletic grouping of flagella-bearing phyla, we found multiple apparent points of origin for flagellar systems on the phylogenetic tree (Figure 2). This highlights a fundamental problem with any simple model of flagellar divergence: although there is some agreement as to the existence of bacterial phyla, there is no consensus on the order of their divergence.
The problem here is that flagella do not fit into the nice, neat nested hierarchy that you’d expect from common ancestry. Quite the opposite — their diversity conflicts with what you would expect from a Darwinian origin of the flagellum. Indeed, the caption for the figure above from the paper states, “Arrows indicate apparent points of origins for flagellar lineages.” Common descent predicts there should be just one arrow, but as you can see on the diagram there are five arrows, because there are no fewer than five clades — widely separated on the tree — that have flagella. This is not what common descent predicts. Common design, on the other hand would predict that complex features like flagella might be re-used in a manner that doesn’t match a nested hierarchy, which is exactly what we see here. Ironically, the nature of bacterial flagellar diversity — far from being a problem for intelligent design — is actually a significant problem for Darwinian evolution.
Baker’s argument here is ironic. We constantly see evolutionists arguing that homology (i.e., sequence similarity) between proteins is evidence that they evolved. In particular, we see this argument made regarding the flagellum. In fact, in a striking comment that appears after his article, Baker himself states that he feels no obligation to provide anything like a stepwise explanation for the evolution of the flagellum precisely because “commonality” between proteins is sufficient to demonstrate flagellar evolution:
I don’t think we need to actually directly show an injectisome can be step by step evolved into a flagellum, the field of experimental evolution is very young! We can look, with greater and greater ease, at the historical genetic record, and look for commonality. This is the basis of most evolutionary genetics. By phylogenetics and common ancestors, we can show that systems share elements, and we do.
Yet in his article he affirmatively quotes Pallen and Matzke saying that the “highly diverse” nature of flagellar proteins also demonstrates their evolution. So now apparently both similarity between proteins and differences between proteins demonstrates evolution. No matter what you find, it demonstrates evolution!
What a sweet deal it must be to be a Darwinian evolutionist. You don’t have to provide any evidence that your theory is true (i.e., offer some semblance of a stepwise evolutionary explanation), and no matter what evidence you do find, it is guaranteed to show that your theory is true!
Indeed, Pallen and Matzke’s paper presents other related contradictions. They repeatedly denigrate “typological” thinking, stating:
As the great evolutionist Ernst Mayr noted, one of Darwin’s greatest achievements was to abolish typological or essentialist thinking from biology; instead, the emphasis in biology is on variation and individuality. Therefore, when discussing flagellar evolution it is important to appreciate that there is no such thing as ‘the’ bacterial flagellum. Instead, there are myriad different bacterial flagella, showing extensive variation in form and function.
Since their article is an explicit attack on ID, I suppose their point is that they’re trying to tag ID as a form of typological thinking, wrongly suggesting that ID can’t accommodate the diversity among flagella. Ironically, however, Pallen and Matkze later admit that “all (bacterial) flagella share a conserved core set of proteins,” numbering around 20 proteins, and they further concede that there is a common core of subsystems found in known bacterial flagella:
Three modular molecular devices are at the heart of the bacterial flagellum: the rotor-stator that powers flagellar rotation, the chemotaxis apparatus that mediates changes in the direction of motion and the T3SS that mediates export of the axial components of the flagellum.
There you have it: despite all the apparent “diversity” of flagella, and the evolutionists’ distaste for “typological thinking,” they admit that all bacterial flagella share a conserved core of about 20 proteins, and a common core (what I would call an irreducible core) of subsystems: a motor, a chemotaxis mechanism, and a secretion apparatus. It seems like the many diverse types of flagella are variations on a common thematic archetype.
And how do we know this core is irreducibly complex? Because the experimental data shows it is. Scott Minnich’s genetic knockout experiments on the E. coli flagellum have shown that it fails to assemble or function properly if any one of its approximately 35 structural parts are missing.
Baker discloses none of this, but simply asserts that the irreducible complexity of the flagellum has been refuted:
Typically, intelligent design proponents persevere despite this evidence. They simply adjust their goal posts by selecting other systems to act as poster boys for irreducible complexity. It is difficult to respond to these movable challenges. But as we learn more about the origins of these and other complex systems, we can at least reduce the number of available candidates used to prop up the theory of intelligent design.
Actually ID proponents haven’t abandoned arguing for the irreducible complexity of the flagellum because, as we’ve seen, it was never refuted in the first place. Rather than moving goal posts, we’re building a formidable team, as ID advocates have expanded their arguments far beyond irreducible complexity. But I suspect that Dr. Baker doesn’t pay much attention to any of that.
Thus, he closes with a typically inaccurate rant about the dangers of intelligent design:
While all this may seem relatively harmless, the intelligent design movement is well funded, slickly presented, and actively challenges educational curricula in many countries. It is a dangerously well-articulated distraction from the large body of evidence supporting evolutionary theory.
Actually our funding is nothing compared to the wealth of support available to upholders of Darwinian evolution. Setting that aside, consider Baker’s claim that ID distracts from the “large body of evidence supporting evolutionary theory.” In fact, as we’ve seen, both evidence and logic contradict Baker’s arguments. In fact, he openly refuses to demonstrate what Darwinian evolution requires: a stepwise evolutionary explanation of the flagellum! He apparently wants to blame ID for his failure to make a convincing case.
The world is very different from the one that many Darwin advocates believe they live in. Despite their protests to the contrary, this debate is far from over.
